Enabling Technologies
Camcore’s primary objectives are Gene Conservation and Tree Improvement, but another critical objective can be summarized as Development of Enabling Technologies. We do research on new methodologies, tools, and techniques to help make tree breeding programs and plantation forestry more efficient and productive. These technologies can be used in both Camcore breeding and testing programs, but can also be used by our members in in their own internal breeding programs. Over the past 10 years, much of this work has focused on ways to assess wood properties. Wood property traits are very important in determining final product quality, but typically are time consuming and expensive to measure on large populations of tree. Our work is focused on finding methods to assess wood properties that are fast, precise, and cheap. Some examples of our projects in this area listed below.
Imaging technologies and Lidar: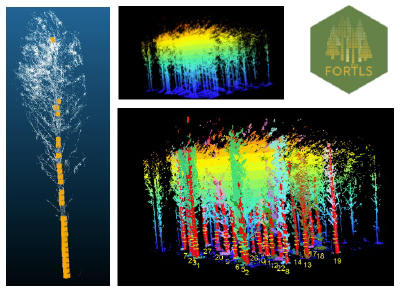
Recently, we have been studying the potential use of new imaging technologies for genetic trial assessment. Many forestry companies around the world are investigating or using aerial photogrammetry and airborne lidar as tools for forest inventory, and in some cases, to measure tree height in research trials. Close-range LiDAR technologies such as Terrestrial Laser Scanner (TLS) and Mobile Laser Scanner (MLS) have also been used in forestry to estimate quickly and accurately stand-level variables for forest management (e.g., timber volume). In addition, they can be used in other research endeavors such us fitting allometric equations for wood volume without destructive sampling, estimation of biomass, etc. Our interest in Camcore is more toward the use of TLS taken from ground level to measure tree diameters, stem straightness, stem taper, branch characteristics, etc., and then to use that information to understand the genetic control of important traits. For that purpose, we have scanned two progeny tests in Eastern North Carolina, USA and we are currently collaborating with experienced colleagues in Spain, Czech Republic and Brazil for algorithm testing and data analysis. To facilitate transferring of results to our members, we are using open-source software for this project and we have tested many R packages available such as, LidR, TreeLS. Currently, we are working closely with the developer of FORTLS (https://molina-valero.github.io/FORTLS/ ), an R package created at the University of Santiago de Compostela in Spain.
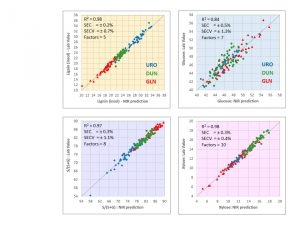
NIR Models to Predict Wood Chemistry
We have done a great deal of work developing NIR (Near Infrared Spectroscopy) models to predict wood chemistry. NIR spectroscopy involves beaming light from many hundreds of wavelengths onto a sample and measuring the reflectance. These data are then combined in multivariate statistical model to predict a chemical trait of interest. Wood chemistry (e.g., sugar and lignin content and composition) has a huge effect on pulp yields and is therefore of critical economic importance in pulp and paper mills.
We have developed global NIR models for both pines and eucalypt wood chemistry. These models are based on hundreds of samples from multiple species and multiple environments, so they can be used to make predictions for wood samples of any age, for any pine or eucalypt species, from any part of the world. These models are highly precise (for any given trait, we typically obtain R2 from 0.85 to 0.95), so the predicted chemical values can be used with confidence to rank families and individual trees for selection.
Handheld NIR Applications
Most NIR work is done with very precise laboratory spectrometers. In recent years, portable handheld NIR machines have been developed. These machines are typically much less expensive than laboratory machines, and offer the advantage of portability. However, these machines usually measure reflectance at many fewer wavelengths than laboratory machines. Camcore has been investigating the utility of a particular handheld NIR machine (ThermoScientific microPhazir) to develop the same kind of wood chemistry models as described above. The resulting models are not quite as precise as from laboratory machines, but typically have R2 from 0.80 to 0.90), so they are still quite useful in a selection and breeding program. We have tested the handheld NIR on both woodmeal and solid wood samples, and obtain good models with both types of sample.
We are also investigating the use of the handheld NIR to measure nutrient content in live green teak foliage. The idea is see if we can evaluate foliage nutrient content in a nursery situation so as to be able to respond quickly to nutrient deficiencies. This work is ongoing.

Resistograph and TreeSonic
Solid wood strength is affected both by wood density and microfibril angle (MFA), the structure of the cellulose microfibers in the cell walls. We are using two tools to measure these traits in standing trees: the IML Resistograph to measure wood density profiles, and the Fakopp TreeSonic to measure acoustic wave velocity, which is highly correlated to MFA. We are using both of these tools to compare species, families and genotypes, to evaluate the genetic control and genetic variation of these traits in different species.
Genomic Selection and Pedigree Reconstruction
Finally, we are investigating the potential of using new molecular marker techniques in operation breeding programs, both Camcore’s and our members’ internal programs. Genomic selection involves the use tens of thousands of molecular markers to assess the genetic value of a tree for specific traits (such as volume growth, disease tolerance, or pulp yield) from the DNA data alone. If this is possible, it will allow breeders to greatly reduce the costs and size of genetic tests in the field.
It may also be possible to use this same type of DNA marker data to “jump-start” a tree breeding program, to save 10 to 20 years at the initiation of a breeding program. Traditionally, tree breeding programs are initiated by making mass selections in natural stands, or for exotic plantation species, in commercial plantations. This is followed by grafting, seed collection, test establishment, and then waiting a number of years for data from those tests. The time required for this first phase of a breeding program could easily be 10 to 20 years, depending on the species. DNA marker data can be used to reconstruct the pedigree of a plantation population, that is to to determine the genealogical relationships among a population of individuals. Effectively, this would allow a breeder to recover “progeny-test-like” data from existing plantations, and to predict breeding values and identify genetically superior individuals at the initial stage of a breeding program.

